Terbium »
PDB 1m9s-6xym »
3ltq »
Terbium in PDB 3ltq: Structure of Interleukin 1B Solved By Sad Using An Inserted Lanthanide Binding Tag
Protein crystallography data
The structure of Structure of Interleukin 1B Solved By Sad Using An Inserted Lanthanide Binding Tag, PDB code: 3ltq
was solved by
E.Peisach,
K.N.Allen,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 30.43 / 2.10 |
| Space group | P 63 2 2 |
| Cell size a, b, c (Å), α, β, γ (°) | 120.584, 120.584, 74.899, 90.00, 90.00, 120.00 |
| R / Rfree (%) | 19.2 / 22.1 |
Terbium Binding Sites:
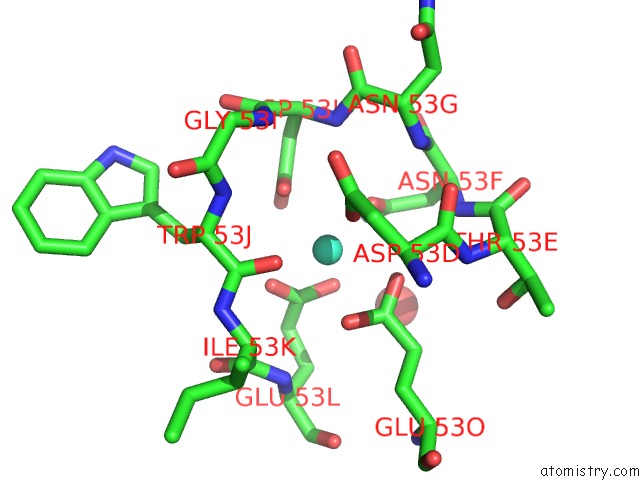
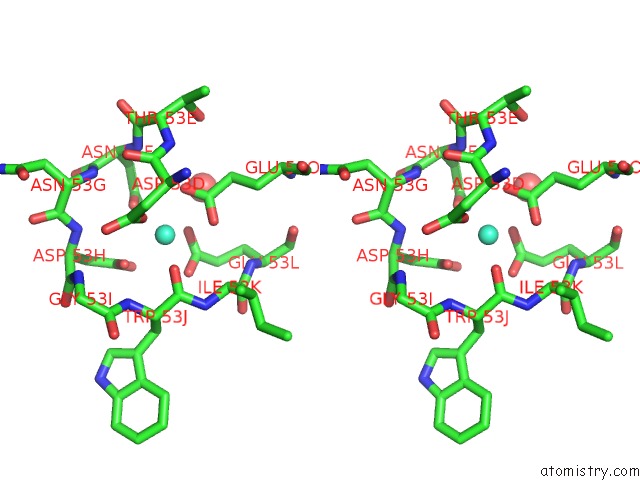
The binding sites of Terbium atom in the Structure of Interleukin 1B Solved By Sad Using An Inserted Lanthanide Binding Tag
(pdb code 3ltq). This binding sites where shown within
5.0 Angstroms radius around Terbium atom.
In total only one binding site of Terbium was determined in the Structure of Interleukin 1B Solved By Sad Using An Inserted Lanthanide Binding Tag, PDB code: 3ltq:
In total only one binding site of Terbium was determined in the Structure of Interleukin 1B Solved By Sad Using An Inserted Lanthanide Binding Tag, PDB code: 3ltq:
Terbium binding site 1 out of 1 in 3ltq
Go back to
Terbium binding site 1 out
of 1 in the Structure of Interleukin 1B Solved By Sad Using An Inserted Lanthanide Binding Tag
Mono view
Stereo pair view

Mono view

Stereo pair view
A full contact list of Terbium with other atoms in the Tb binding
site number 1 of Structure of Interleukin 1B Solved By Sad Using An Inserted Lanthanide Binding Tag within 5.0Å range:
|
Reference:
K.Barthelmes,
A.M.Reynolds,
E.Peisach,
H.R.Jonker,
N.J.Denunzio,
K.N.Allen,
B.Imperiali,
H.Schwalbe.
Engineering Encodable Lanthanide-Binding Tags Into Loop Regions of Proteins. J.Am.Chem.Soc. V. 133 808 2011.
ISSN: ISSN 0002-7863
PubMed: 21182275
DOI: 10.1021/JA104983T
Page generated: Tue Aug 19 05:58:14 2025
ISSN: ISSN 0002-7863
PubMed: 21182275
DOI: 10.1021/JA104983T
Last articles
W in 1DV4W in 1FR3
W in 1GUG
W in 1H9R
W in 1H9K
W in 1H0H
W in 1FEZ
W in 1FKA
W in 1E3P
W in 1E18