Terbium »
PDB 1m9s-6xym »
6rqa »
Terbium in PDB 6rqa: Crystal Structure of the Iminosuccinate Reductase of Paracoccus Denitrificans in Complex with Nad+
Protein crystallography data
The structure of Crystal Structure of the Iminosuccinate Reductase of Paracoccus Denitrificans in Complex with Nad+, PDB code: 6rqa
was solved by
J.Zarzycki,
F.Severi,
L.Schada Von Borzyskowski,
T.J.Erb,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 29.40 / 2.56 |
| Space group | P 21 21 21 |
| Cell size a, b, c (Å), α, β, γ (°) | 50.386, 72.407, 164.266, 90.00, 90.00, 90.00 |
| R / Rfree (%) | 17.6 / 22.5 |
Terbium Binding Sites:
The binding sites of Terbium atom in the Crystal Structure of the Iminosuccinate Reductase of Paracoccus Denitrificans in Complex with Nad+
(pdb code 6rqa). This binding sites where shown within
5.0 Angstroms radius around Terbium atom.
In total 3 binding sites of Terbium where determined in the Crystal Structure of the Iminosuccinate Reductase of Paracoccus Denitrificans in Complex with Nad+, PDB code: 6rqa:
Jump to Terbium binding site number: 1; 2; 3;
In total 3 binding sites of Terbium where determined in the Crystal Structure of the Iminosuccinate Reductase of Paracoccus Denitrificans in Complex with Nad+, PDB code: 6rqa:
Jump to Terbium binding site number: 1; 2; 3;
Terbium binding site 1 out of 3 in 6rqa
Go back to
Terbium binding site 1 out
of 3 in the Crystal Structure of the Iminosuccinate Reductase of Paracoccus Denitrificans in Complex with Nad+
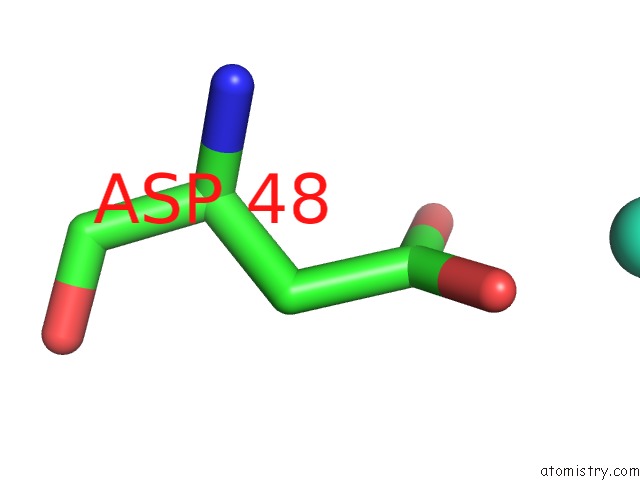
Mono view
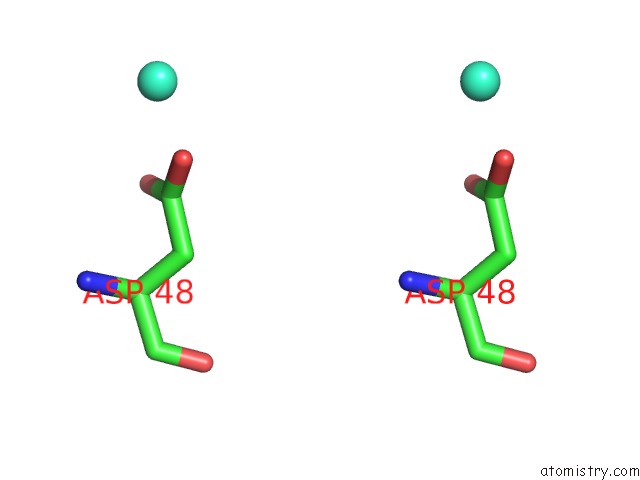
Stereo pair view

Mono view

Stereo pair view
A full contact list of Terbium with other atoms in the Tb binding
site number 1 of Crystal Structure of the Iminosuccinate Reductase of Paracoccus Denitrificans in Complex with Nad+ within 5.0Å range:
|
Terbium binding site 2 out of 3 in 6rqa
Go back to
Terbium binding site 2 out
of 3 in the Crystal Structure of the Iminosuccinate Reductase of Paracoccus Denitrificans in Complex with Nad+
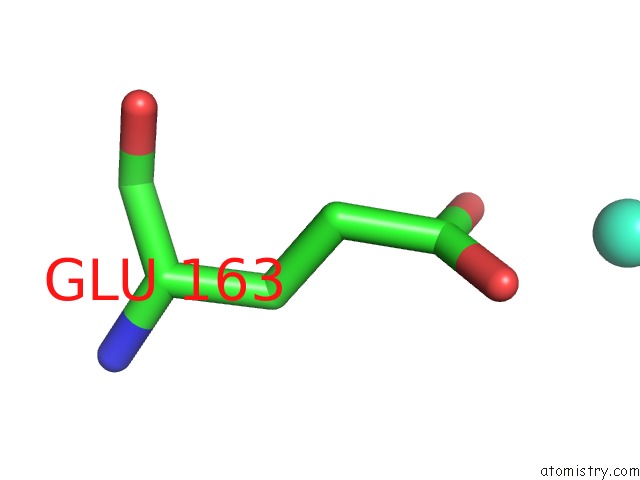
Mono view
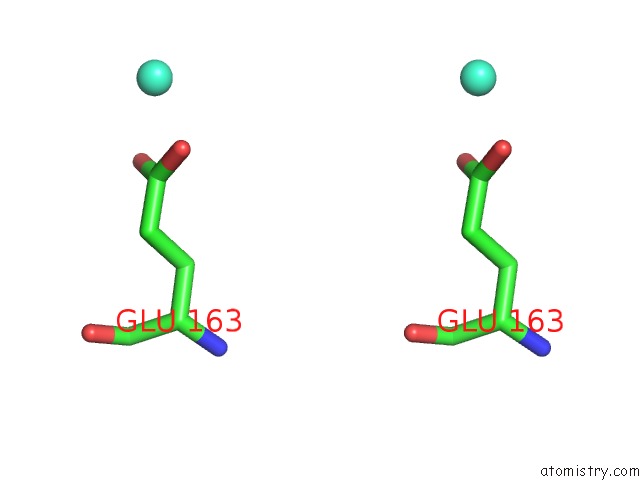
Stereo pair view

Mono view

Stereo pair view
A full contact list of Terbium with other atoms in the Tb binding
site number 2 of Crystal Structure of the Iminosuccinate Reductase of Paracoccus Denitrificans in Complex with Nad+ within 5.0Å range:
|
Terbium binding site 3 out of 3 in 6rqa
Go back to
Terbium binding site 3 out
of 3 in the Crystal Structure of the Iminosuccinate Reductase of Paracoccus Denitrificans in Complex with Nad+
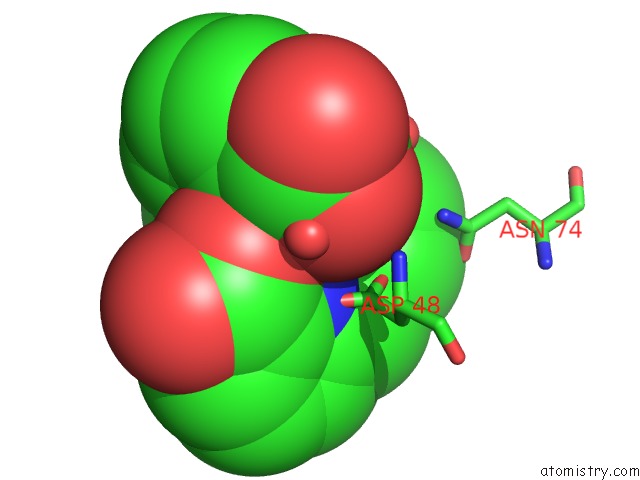
Mono view
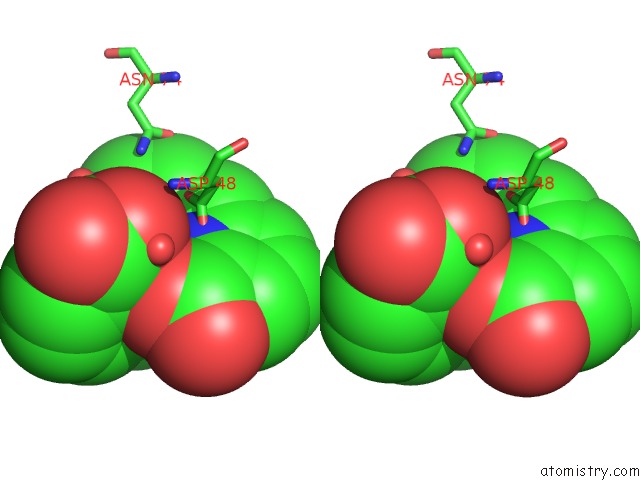
Stereo pair view

Mono view

Stereo pair view
A full contact list of Terbium with other atoms in the Tb binding
site number 3 of Crystal Structure of the Iminosuccinate Reductase of Paracoccus Denitrificans in Complex with Nad+ within 5.0Å range:
|
Reference:
L.Schada Von Borzyskowski,
F.Severi,
K.Kruger,
L.Hermann,
A.Gilardet,
F.Sippel,
B.Pommerenke,
P.Claus,
N.S.Cortina,
T.Glatter,
S.Zauner,
J.Zarzycki,
B.M.Fuchs,
E.Bremer,
U.G.Maier,
R.I.Amann,
T.J.Erb.
Marine Proteobacteria Metabolize Glycolate Via the Beta-Hydroxyaspartate Cycle. Nature V. 575 500 2019.
ISSN: ESSN 1476-4687
PubMed: 31723261
DOI: 10.1038/S41586-019-1748-4
Page generated: Fri Oct 11 08:40:53 2024
ISSN: ESSN 1476-4687
PubMed: 31723261
DOI: 10.1038/S41586-019-1748-4
Last articles
Cl in 5RAECl in 5RAD
Cl in 5RAC
Cl in 5RAB
Cl in 5RA9
Cl in 5RAA
Cl in 5RA8
Cl in 5RA7
Cl in 5RA4
Cl in 5RA6